Sample Submission Guide
We recommend users to QC their DNA samples prior to shipment and attaching the results (gel, trace analysis, concentration and ratios) to the sample submission form. Please review our guidelines to make sure your sample is of sufficient quality to be sequenced. The table below contains the required sample features for the different applications.
Minimum requirements for incoming samples
| Application | Optimal input DNA/RNA amount | Minimum concentration | maximum volume at the minimum sample concentration | Sample buffer | Comments |
|---|---|---|---|---|---|
| WGS HiFi | >5 ug, better >8 ug | ideal: 110 ng/µl minimum: 40 ng/µl |
46 µl | low TE (10mM Tris-HCl & 0.1mM EDTA) | For multiple SMRT cells, higher amounts are required. The majority of DNA mass must be above 20 kb (measured at Femto Pulse or Fragment Analyzer). |
| WGS HiFi Low Input | 500-3000 ng | 11 ng/µl | 46 µl | low TE (10mM Tris-HCl & 0.1mM EDTA) | The majority of DNA mass must be above 10 kb (measured at Femto Pulse or Fragment Analyzer). |
| Amplicon up to 1 kb | 1,000 ng | 25 ng/µl | 40 µl | low TE (10mM Tris-HCl & 0.1mM EDTA) | Shipped as purified amplicons. Includes extra sample for accidental sample loss during library prep. |
| Amplicon >1 kb | >1,500 ng | 25 ng/µl | 40 µl | low TE (10mM Tris-HCl & 0.1mM EDTA) | Shipped as purified amplicons. Includes extra sample for accidental sample loss during library prep. |
| Iso-Seq | 1,000 ng total RNA | 43 ng/µl | 7 µl | nuclease-free water | Total RNA shipped in dry ice, RIN >=7. Includes extra sample for additional steps (e.g. rRNA depletion) and accidental sample loss during library prep. |
| PacBio 16S | 5 ng | 2 ng/µl | 15 µl | low TE (10mM Tris-HCl & 0.1mM EDTA) or water | To serve as template for PCR. Adopting the PacBio protocol, amplifying the V1-V9 region, ~1.5 kb. 260/280 at ~1.8 |
DNA sample requirements
Essential requirements of your DNA sample:
- is double-stranded DNA, as single-stranded DNA is not compatible with the library preparation process.
- has not undergone multiple freeze-thaw cycles as they can lead to DNA damage.
- has not been exposed to high temperatures (e.g. > 37 °C for 1 hour), pH extremes (< 6 or > 9). Air drying of pellets is preferred to overheat drying. Avoid the over drying of genomic DNA. Do not heat when drying in a speed-vacuum.
- has an OD 260/280 nm ratio of 1.8 to 2.0 (measured at Nanodrop).
- does not contain insoluble material.
- does not contain RNA.
- has not been exposed to intercalating fluorescent dyes or ultraviolet radiation.
- does not contain chelating agents (e.g. EDTA), divalent metal cations (e.g. Mg2+), denaturants (e.g. guanidinium salts, phenol), or detergents ( e.g. SDS, Triton-X100).
- does not contain carryover contamination from the starting organism/tissue (heme, humic acid, polyphenols, polysaccharides, etc.).
Sample concentration should be 100 ng/μl
or higher, the amount of the DNA should be at least 8 μg (for a single SMRT cell project).
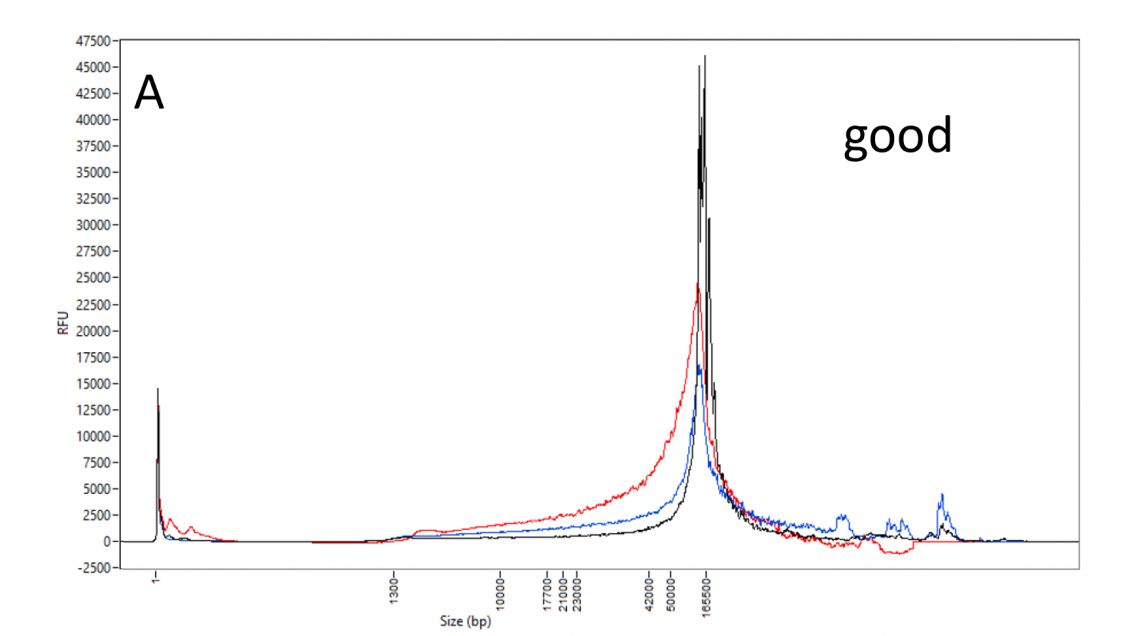
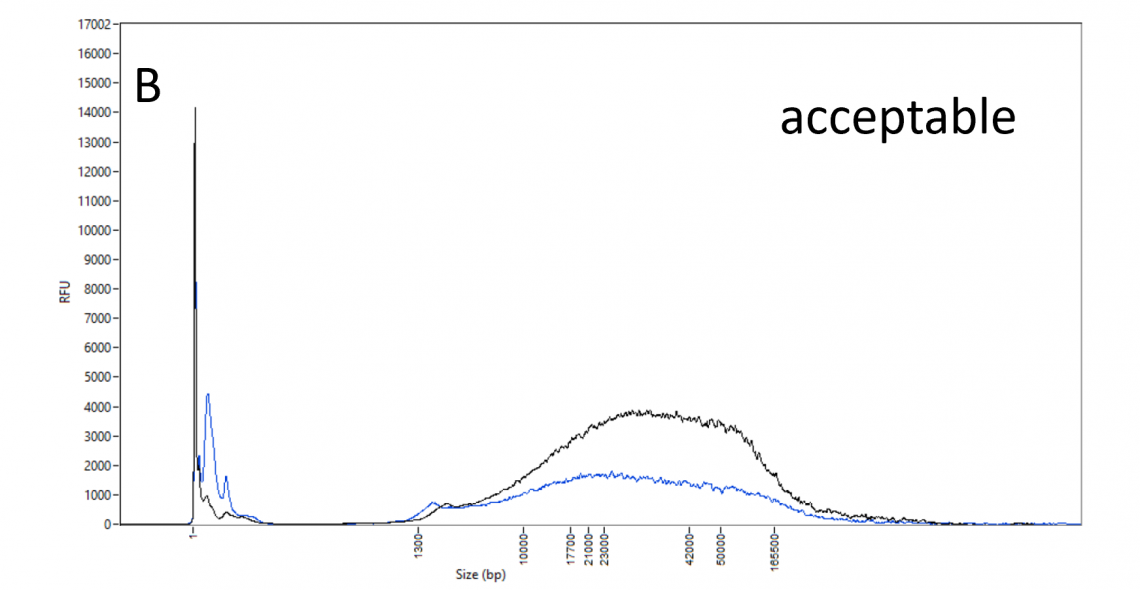
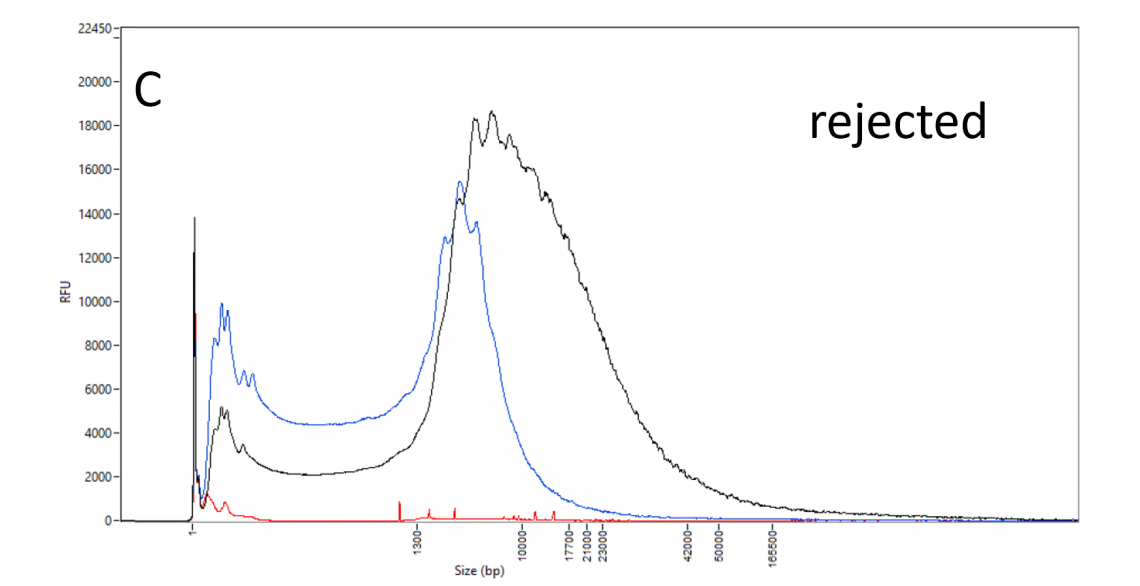
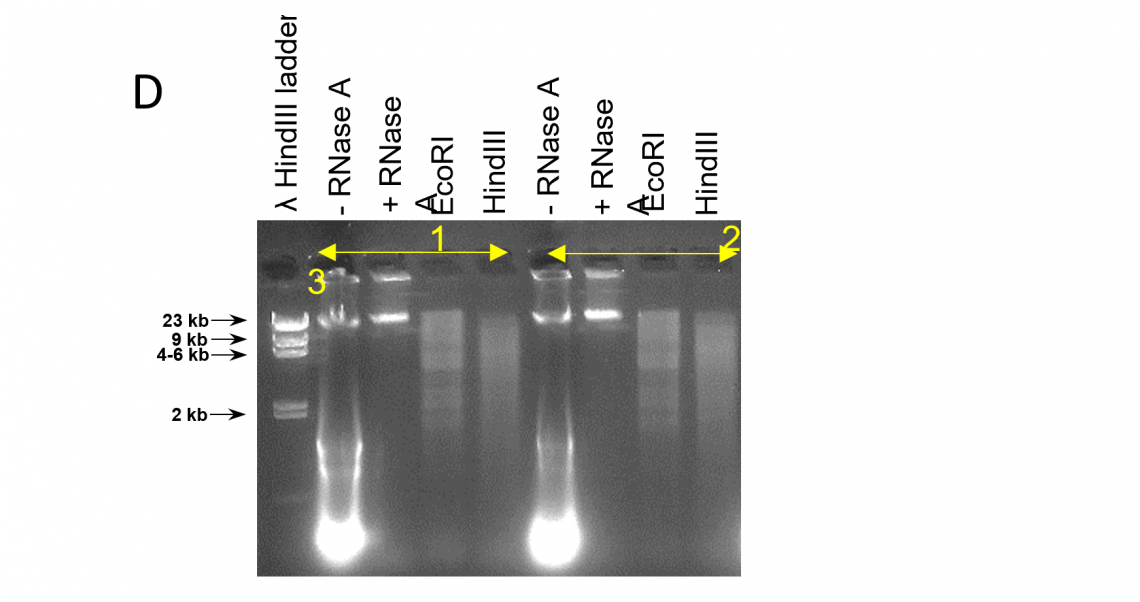
DNA size profile and restriction enzyme accessibility:
DNA_QC_features_220630-A.png
DNA_QC_features_220630-B.png
DNA_QC_features_220630-C.png
DNA_QC_features_220630-D.png
RNA sample requirements
RNA samples should have a RNA integrity number (RIN) of 7 or more. Ideally, samples should have a RIN > 8.
Samples with RIN < 7 can still be processed, but the risk of significant underperformance or even failure is greater. All RNA samples should be shipped in dry ice.
Sequencing of PCR amplicons
Products of PCR should be shipped upon in-house bead purification, in EB buffer and in a volume no larger than 47 µl.
The DNA amount varies depending on the amplicon size, and is reported in page 3 of this protocol.
Samples should be shipped in ice blocks or at a lower temperature.
16S amplification and sequencing
Samples should be arrayed in plates (if less than 16, single tubes are also accepted), preferably normalized at 2-10 ng/µl, in water or low TE buffer.

